New Technologies Can Step Up Fight Against Antimicrobial Resistance and Sepsis
Sepsis and antimicrobial resistance (AMR) account for growing numbers of deaths and healthcare costs worldwide. It is believed that sepsis causes over 5 million deaths annually. One report predicts that AMR could cause over 10 million deaths globally per year by 2050.
Both sepsis and AMR in the clinic can be challenging. While good antibiotic stewardship promotes careful diagnostic testing and fewer antibiotic prescriptions, sepsis guidelines recommend broad-spectrum antibiotics early in suspected bacterial infection. Additionally, there is a fundamental diagnostic problem: When a patient presents with signs and symptoms of infection, there are few tools available to the physician to quickly and accurately guide antibiotic selection. Emergency room physicians struggle with decision-making in many of the approximately 15 million annual patients with suspected acute infection and sepsis.
There are inherent drawbacks with diagnostics for acute infections that typically seek to either identify pathogens directly or to measure specific host biomarkers to establish a diagnosis indirectly:
- Pathogen testing is typically slow
- With the exception of newer respiratory pathogen tests, most locations in the body are not easily sampled (e.g., there is no easy way to sample a belly infection at the bedside).
- Most patients with infections do not have pathogens that can be detected in the blood, so a negative result does not rule out the need for antibiotics.
For instance, a patient in the ED with radiographically confirmed pneumonia might get a nasal respiratory pathogen panel, sputum culture, blood culture, and urine antigen tests. With all of that testing, 62% of the time no pathogen will be found (https://doi.org/10.1056/NEJMoa1500245).
Biomarker testing is typically faster, but common biomarkers such as C-reactive protein (CRP) and procalcitonin are neither specific for infection nor accurate enough to be routinely used for antibiotic prescribing. Thus, in acute settings physicians must decide whether to initiate antibiotics without confirmatory testing, typically choosing antibiotics based on guidelines and local antibiograms rather than microbiological data. Data suggest that physicians frequently guess wrong, both in choosing if to prescribe and what to prescribe for optimal treatment.
The need for more accurate diagnostics for acute infections is clear. New advances in acute infection diagnosis are creating opportunities to ensure that patients get the right treatments in a cost-efficient manner, a workflow that will be critical to tackling AMR and sepsis.
Broadly, new approaches for acute infection diagnosis can be split into three categories
- Rapid pathogen detection
- Rapid pathogen antibiotic susceptibility testing (AST)
- Host-response measurements
Some technologies are combining rapid pathogen detection and rapid AST, typically genotypic (detection of resistance markers) not phenotypic (direct measurement of bacterial growth inhibition by antibiotics). Elsewhere there are excellent reviews of rapid ID and AST and host response technologies.
To understand the role that these approaches may play in antibiotic prescribing, it is important to consider the decisions that physicians must make for patients with suspected infection:
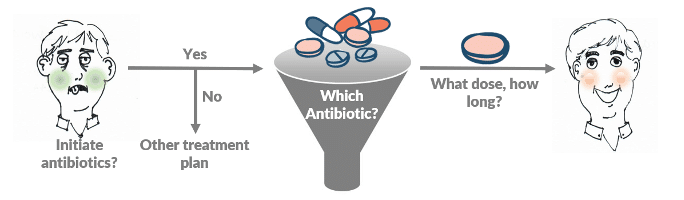
Physicians may use pathogen-detection and host-response technologies at all phases, but the primary utility of host-response is in choosing whether to start antibiotics and when to stop them, while the primary utility of pathogen-detection (and especially AST) is in choosing the right antibiotics for the patient. (Novel host-response technologies that can rapidly and more accurately identify bacterial and viral infections may show improved utility for both starting and stopping antibiotics but are still in development. Inflammatix’s tests fall into this category.)
Taking into account turnaround time and clinical impact, future diagnostic workflows may differ dramatically from today’s practice. In the future, perhaps testing will start with a rapid (<30 minute) host-response test that identifies the infection as viral or bacterial (aiding in the prescribing decision). Host-response indicators that suggest a bacterial infection would be followed up with immediate initial antibiotics and then rapid (1-6 hour) direct pathogen detection and AST, so that the patient may only get one broad-spectrum dose before being narrowed to appropriate therapy. Host-response monitoring could then help with the decision on when to stop (or switch to orals and discharge). In occasional severe or chronic infections, follow-up testing with metagenomics for pathogen nucleic acids (hours to days) may aid in identifying rare infections.
Patients with bacterial infections will benefit from early, accurate antibiotic administration, and this will aid in reducing morbidity and mortality from sepsis. Avoiding unnecessary antibiotics in patients without bacterial infections will decrease antibiotic-associated adverse drug events, reduce costs, and reduce AMR. A combination of rapid host-response profiling and AST should be just what the doctor ordered.
Timothy E. Sweeney, M.D., Ph.D., is cofounder and chief executive officer of Inflammatix, Inc. Inflammatix is a molecular diagnostics company building rapid, novel host-response tests for acute infectious and inflammatory conditions. Dr. Sweeney trained in general surgery at Stanford before cofounding Inflammatix. He can be reached at [email protected].