By Kirindi Choi, Ljubomir Buturovic, Roland Luethy, Inflammatix, Inc.
Introduction
Recently, generative artificial intelligence (AI) models for text, images, and video have made major progress and achieved worldwide attention among experts and the public, including initial applications in medicine [1]. The application of generative AI to problems in genomics (the study of genes and their functions) has, understandably, been less visible, but has nevertheless important applications. In this blog, we assess several open-source and commercial tools which can be used to generate high-quality genomic data and discuss potential applications. We focus on transcriptomic applications (a subfield of genomics), with tabular data representing gene expression (the measurements of abundance of gene products in cells). Transcriptomics has significant applications in bioinformatics research and increasingly in clinical care as a new class of diagnostics and prognostics [2-5].
One of the main use cases for synthetic genomic and transcriptomic data is sharing data while preserving privacy. Some ideas include the following: an organization may wish to organize a Kaggle competition for its transcriptomic problem by using synthetic data based on patient data, thereby preserving the privacy of the patient data; or an organization may send synthetic data to a software vendor to report and reproduce a bug, again without sending sensitive data.
Another potential use case is to improve classification accuracy by adding synthetic tabular data to the training set for Machine Learning (ML) models. However, in the available literature, we have not seen convincing evidence of this approach being successful. Thus, we believe that this use case remains hypothetical.
In this blog, we compare the quality of the transcriptomic synthetic data created using different generative AI tools.
Methods
We selected and compared two open-source and two commercial synthetic data generators (available through Python API).
- We selected the following open-source Python tools:
- CTGAN (conditional tabular generative adversarial network) [6] from SDV (Synthetic Data Vault) [7] and
- Gaussian Copula (Gaussian Multivariate) [8] in SDV Copulas library.
- We selected the following commercial solutions:
- LSTM and ACTGAN (an alternate implementation of CTGAN) cloud-based APIs from Gretel.ai ([9], [12], [13], [14]).
- To evaluate quality of the synthesized data, we used two different metrics:
- SDMetrics (synthetic data metrics) [10] from SDV.
- Cross-validation AUROC (Area Under Receiver Operating Characteristic) of a binary logistic regression classifier trained on real (positive class) and corresponding synthetic (negative class) data. The idea is that high-quality synthetic data should be difficult to discriminate (classify) from real data, therefore such data should have an AUROC of approximately 0.5. This quality metric has a low false-negative rate: data which fail the metric are unlikely to be high-quality. However, data with AUROC close to 0.5 may still not be high quality
Per [11], we also applied duplicate detection steps after the data were synthesized: we detected and discarded any duplicates within the synthesized dataset, and detected and discarded any synthesized data that were replicates of the real data.
For the data synthesis, we used default values for the parameter settings of the data synthesizers except batch size and number of epochs (Table 1).
We used a real data set with 9,654 patient samples and used the expression levels of 29 genes for each sample [5]. We synthesized 6 sets of data: 4 datasets using SDV tool, and 2 datasets using Gretel tool. The synthetic datasets had 1,000 samples each. The SDV datasets were created on the same EC2 instance with four vCPUs, whereas Gretel.ai is a cloud-based service. Since ACTGAN is a variant of CTGAN, we also ran a CTGAN with the same hyper parameter settings as ACTGAN’s default parameters that use larger network than CTGAN’s default parameters.
We computed an AUROC quality metric as follows. For each set of synthesized data, we used the synthesized data and the real data as the training set (consisting of 10,654 samples in total) wherein synthetic data was considered positive class and real data was considered negative class. We then estimated a cross-validation AUROC for the said training set using scikit-learn Logistic Regression model for binary classification and Optuna [15] hyperparameter search. The AUROC reported corresponds to the highest AUROC found by the hyperparameter search. Ideally, a classifier should not be able to distinguish synthetic data from real. Thus, the corresponding cross-validation AUROC should be as close as possible to 0.5.
The second metric was computed using the SDMetrics package. It evaluates the marginal distributions and pairwise trends between columns. Its overall quality score is an average of all metric scores (i.e., KSComplement, TVComplement, CorrelationSimilarity and CategoryCoverage). The score ranges from 0 to 1 with 1 meaning the best quality.
Results
We performed the duplication check on 10,000 synthetic samples and found no duplicates within synthetic data nor copies of real data. Thus, this QC (Quality Control) step may be redundant.
Besides fine-grained and overall scores, SDMetrics offers convenient comparison visualizations including density plot per column between the synthesized data and the real data.
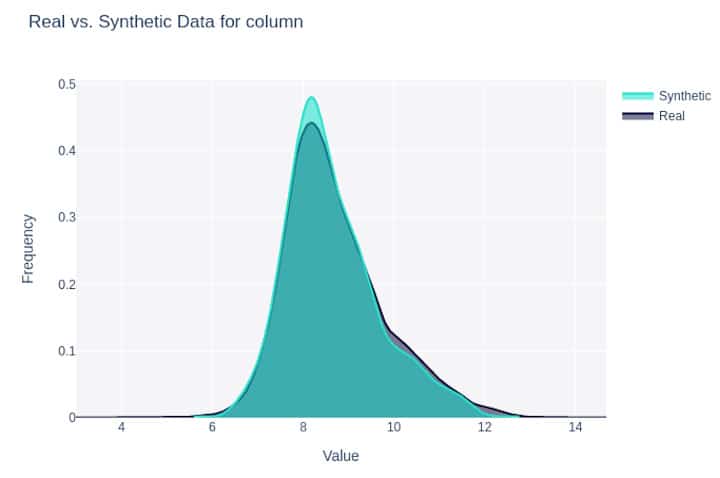
Figure 1: Density plot for a feature synthesized with Gretel LSTM (#6 below).
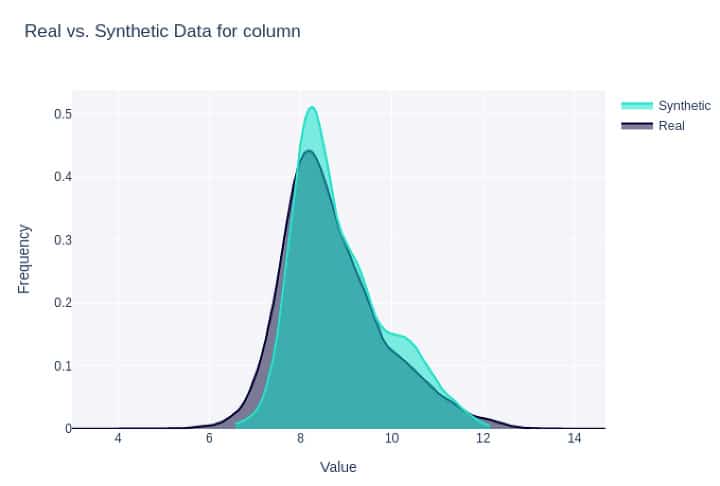
Figure 2: Density plot for the same feature as in Fig. 1 synthesized with CTGAN (#3 below).
All synthetic datasets except Gretel LSTM were easily and highly distinguishable from the real data with AUROCs >= 0.87. For example, for CTGAN with 50 epochs and batch size of 100, a linear classifier could accurately distinguish between real and synthetic data with high accuracy (AUROC = 0.963). Overall, Gretel LSTM performed best in generating data that mimics the set of real data, with an AUROC of 0.623 and the highest SDMetrics overall quality score of 0.949. A distant second was SDV’s CTGAN with 100 epochs and larger dimensions with SDMetrics quality score of 0.927 and was easily distinguishable from real data with AUROC of 0.87. Notably, between the two synthetic datasets generated using Gretel, the LSTM tool was substantially better than their ACTGAN tool.
The AUROC metric proved to be very useful. It was significantly more intuitive and familiar to users than the SDMetric Overall Quality score, yet the rankings of the methods obtained by the two metrics were virtually identical.
| Parameters Overridden | Timing (mins) | AUROC Synthetic vs Real Data | SDMetrics Overall Quality | |
| 1) CTGAN | 50 epochs & 100 batch size | 3.95 | 0.963 | 0.869 |
| 2) CTGAN | 100 epochs & 100 batch size | 7.3 | 0.903 | 0.902 |
| 3) CTGAN | 100 epochs & 100 batch size & ACTGAN’s default parameters | 24.45 | 0.87 | 0.927 |
| 4) Gaussian Copula | defaults | 7.31 | 0.929 | 0.903 |
| 5) Gretel ACTGAN | 50 epochs | 2 | 0.923 | 0.817 |
| 6) Gretel LSTM | 50 epochs | 12.34 | 0.623 | 0.949 |
We observed that turning verbose mode on for CTGAN added excessive amount of time to our data synthesis process and thus we kept verbose off for the runs as shown above.
As expected, we also observed that for the three CTGAN runs with different hyper parameter values, the performance results differed. This suggests some hyperparameter optimization for data synthesizer training can be beneficial for a given set of real data.
In the future we plan to add the Transformer-based NEMO tabular data generator from NVIDIA [11] to our evaluations.
Conclusion
We found that the LSTM synthetic data generator from Gretel.ai is the best among the six solutions that we compared by a wide margin of 0.25 AUROC points between Gretel.ai and the next best software. Interestingly, it is based on LSTM, which to our knowledge has not been widely used for generating synthetic non-temporal tabular data.
Our findings are only based on one internal transcriptomic dataset with numeric features and may not generalize to other data. Nevertheless, we think it is an important data point because the Gretel LSTM was substantially better than any other tool, meaning that it may be inherently superior. We also found the AUROC to be an especially useful and intuitive quality metric in benchmarking performance of these different techniques.
References
- Shah NH, Entwistle D, Pfeffer MA. Creation and Adoption of Large Language Models in Medicine. JAMA. 2023 Aug 7.
- Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, Geyer Jr CE, Dees EC, Goetz MP, Olson Jr JA, Lively T. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. New England Journal of Medicine. 2018 Jul 12;379(2):111-21.
- Alexander EK, Kennedy GC, Baloch ZW, Cibas ES, Chudova D, Diggans J, Friedman L, Kloos RT, LiVolsi VA, Mandel SJ, Raab SS. Preoperative diagnosis of benign thyroid nodules with indeterminate cytology. New England Journal of Medicine. 2012 Aug 23;367(8):705-15.
- Pham MX, Teuteberg JJ, Kfoury AG, Starling RC, Deng MC, Cappola TP, Kao A, Anderson AS, Cotts WG, Ewald GA, Baran DA. Gene-expression profiling for rejection surveillance after cardiac transplantation. New England Journal of Medicine. 2010 May 20;362(20):1890-900.
- Brakenridge SC, Chen UI, Loftus T, Ungaro R, Dirain M, Kerr A, Zhong L, Bacher R, Starostik P, Ghita G, Midic U. Evaluation of a multivalent transcriptomic metric for diagnosing surgical sepsis and estimating mortality among critically ill patients. JAMA Network Open. 2022 Jul 1;5(7):e2221520-.
- CTGAN (conditional tabular generative adversarial network). https://github.com/sdv-dev/CTGAN
- SDV (Synthetic Data Vault). https://github.com/sdv-dev
- Gaussian Copula (GaussianMultivariate). https://github.com/sdv-dev/Copulas
- LSTM and ACTGAN cloud-based APIs. https://gretel.ai/
- SDMetrics (synthetic data metrics). https://github.com/sdv-dev/SDMetrics
- Synthetic Tabular Data Generation Using Transformers, March 2023 https://www.nvidia.com/en-us/on-demand/session/gtcspring23-dlit52224/
- Gretel LSTM https://docs.gretel.ai/reference/synthetics/models/gretel-lstm
- Gretel ACTGAN https://docs.gretel.ai/reference/synthetics/models/gretel-actgan
- Gretel blog about ACTGAN https://gretel.ai/blog/scale-synthetic-data-to-millions-of-rows-with-actgan
- Optuna: A hyperparameter optimization framework https://optuna.readthedocs.io/en/stable/index.html
